
Traces in the Water
Using eDNA to Protect California's Ecosystems
Tiny fragments of DNA permeate the air, soil, and water around us. This environmental DNA (eDNA) unlocks a non-invasive way to monitor biodiversity and detect species that might otherwise go unnoticed. Dr. Andrea Schreier, an adjunct associate professor in the Department of Animal Science in the College of Agriculture and Environmental Sciences at UC Davis, began working with eDNA in the 2010s when its potential and limitations were first being explored. Working alongside her team in the Genomic Variation Laboratory (GVL), they are harnessing eDNA to monitor invasive species, protect sensitive ecosystems, and advance conservation efforts across California.
From Water Samples to Species Detection
Despite its many benefits, eDNA is not without challenges, including contamination from water movement and runoff. Water movement, such as tides, currents, or storm runoff, can carry eDNA from distant sources, introducing DNA from species not present in the immediate sampling area. Preventing contamination is critical because even trace amounts of DNA from other sources can compromise the accuracy of results, leading to false positives or obscuring the presence of target species, sometimes with significant consequences. As Dr. Schreier explains, "A false positive could trigger unnecessary and costly actions, while a false negative could allow an invasive species more time to establish and spread undetected." This underscores the importance of expertise in interpreting results and designing targeted assays that minimize errors, ensuring effective management of invasive species.
In the Bay-Delta, turbidity—caused by suspended particles in the water—and bidirectional flow, where water moves in different directions depending on tides or currents, further complicate eDNA interpretation. These conditions make it harder to pinpoint the exact source of the eDNA and increase the likelihood of contamination from unrelated areas, making contamination control even more essential. To mitigate these risks, eDNA water samples are collected in sterile bottles, stored on ice, and transported to a "clean lab" designed specifically to minimize contamination. As Dr. Schreier explains, the lab is “as isolated as the fire marshal would allow,” incorporating UV lighting and weekly bleaching protocols to prevent cross-contamination with other ongoing research.

Fighting Invaders: How eDNA Tracks Nutria, Mussels, and Other Threats
The Genomic Variation Laboratory, in partnership with the California Department of Water Resources (CDWR), focuses on three invasive species in the Bay Delta: Quagga (Dreissena bugensis) and Zebra (Dreissena polymorpha) mussels and the semiaquatic rodent Nutria (Myocastor coypus). Each species presents unique challenges to the Bay Delta ecosystem. The mussels clog critical water transport infrastructure, and once established, physical removal becomes a costly and ongoing task. Beyond this, they can also alter water chemistry, negatively impacting native species. Dr. Schreier points out that zebra mussels filtered so much phytoplankton in the Great Lakes that they drastically changed the water chemistry—raising concerns they could do the same in California’s Bay Delta.
Nutria poses a different kind of threat by burrowing into levees, which increases flood risks. As prolific breeders, their populations are difficult to control. They also prefer swampy environments, where thick, mucky water presents a particular challenge for most eDNA sampling methods, which require the water to pass through a filter. To overcome this challenge, the Genomic Variation Laboratory has been experimenting with new methods to enhance sample collection in these difficult environments. Dr. Emily Funk, an associate specialist in the Department of Animal Science at UC Davis and lab manager of the GVL, has been testing Zymo kits—originally developed for detecting pathogens in wastewater. These kits don’t require filtering, potentially improving the chances of detecting eDNA in nutria habitats without the complications of filtering.

A New Invasive Threat: Golden Mussels and the Role of eDNA
The recent discovery of golden mussels (Limnoperna fortunei) in Northern California's Port of Stockton signals a new invasive threat. Initial identification relied on visual observations and was later confirmed through a genetic analysis led by Dr. Funk. “When they initially sent the specimens, they were unknown mussels. As I was prepping and sequencing the samples, the Smithsonian provided a genus ID, and then, after sequence analysis, we identified the species as golden mussel.” Funk explained, providing the first concrete evidence of the species in North America.
Shortly after the initial detection, water samples collected near the Port of Stockton revealed the presence of golden mussel eDNA, and additional specimens have since been observed. However, tracking golden mussels in the Bay-Delta presents the same challenges as tracking other species using eDNA. “Environmental conditions like turbidity and sunlight impact how long eDNA remains detectable,” explains Dr. Melinda Baerwald, Environmental Program Manager at the CDWR. In response, researchers are working to refine sampling protocols and improve detection methods for challenging environments like the Bay-Delta.
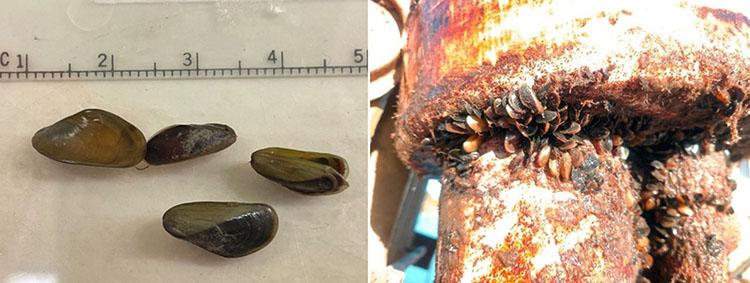
To manage this emerging threat, CDWR and other agencies are expanding eDNA surveillance to additional at-risk areas, particularly around the State Water Project infrastructure. Boat traffic is thought to have been the original carrier of the species into North America, and still poses a significant risk for further spreading of golden mussels, making decontamination protocols essential. As Dr. Baerwald notes, collaborative efforts between multiple organizations will be crucial in determining the mussels' geographic spread. These collaborative efforts aim to standardize sampling protocols and share data, creating a comprehensive strategy to prevent further spread.
Technologies and Techniques on the Horizon
Looking to the future, the GVL is focused on developing field assays that allow for quick detection of eDNA and eRNA without needing a lab, equipping managers with tools to detect nutria and invasive mussels directly from water samples. In addition to supporting field testing, they are working to incorporate eRNA techniques into their repertoire. Unlike eDNA, eRNA degrades very quickly and can only be shed by living organisms, offering real-time insights and helping researchers differentiate between current and past inhabitants of an environment. Dr. Funk offers an example: a boat spending time in a reservoir that contains invasive mussels, then traveling to a clean reservoir may shed eDNA into that water without having mussels attached to it, possibly leading to false positive results that would be far less likely to occur with eRNA.

Preserving Biodiversity: Using eDNA to Protect Endangered and Fragile Species
Beyond invasive species, eDNA can be enlisted in efforts to monitor listed species and those sensitive to handling and disturbance. Dr. Schreier points to Delta smelt (Hypomesus transpacificus), an endangered species of fish found only in the San Francisco estuary, as an example. These small, slender-bodied fish are susceptible to handling-related mortality, and using eDNA to detect their presence can greatly reduce the need for handling.

Ever wonder how scientists teach the next generation about eDNA?
The Genomic Variation Lab hosts a UC Davis Picnic Day activity that has people “fishing” for DNA strands made of color-sequenced beads. The strands are hidden in a tub of rice, creating a fun, hands-on activity in which children can decode their bead strands using a chart and match them to a specific species.
The insights gained from eDNA are not only enhancing conservation science but are also influencing management decisions and habitat protection policies. Working in partnership with the US Fish and Wildlife Service, the GVL is using eDNA to monitor vernal pool communities. Traditional sampling methods like dip netting can be disruptive to these sensitive habitats, and some species, such as vernal pool fairy shrimp and tadpole shrimp, are challenging to differentiate visually without harming them. However, eDNA offers a way to study their populations and habitats with minimal trampling and mortality. This research will likely change how these habitats are monitored long into the future, as Dr. Schreier notes “I'm confident that the work my student (Anderson Tate) is doing will directly inform management actions at the refuges he’s studying.”
The Future of eDNA
eDNA is revolutionizing how scientists monitor ecosystems, offering a non-invasive, precise, and scalable method for detecting species in complex environments. By enabling early detection of invasive species like nutria, zebra mussels, quagga mussels and now golden mussels, eDNA empowers researchers and resource managers to respond quickly and mitigate potential ecological damage.
Looking ahead, the integration of advanced techniques like eRNA and field-based detection methods promises to expand the applications of eDNA, making it a critical tool for conservation efforts and invasive species management. As Dr. Schreier’s work at the Genomic Variation Laboratory demonstrates, eDNA is not just a scientific innovation—it’s a transformative approach to preserving biodiversity and protecting California’s ecosystems.
